👋DefenseFinder news !
Thinking of how we can better help the community, we provide a new website that includes a wiki on defense systems, a structure database and precomputed DefenseFinder results on >22k genomes.
https://defensefinder.mdmlab.fr
Here is the preprint : https://www.biorxiv.org/content/10.1101/2024.01.25.577194v1
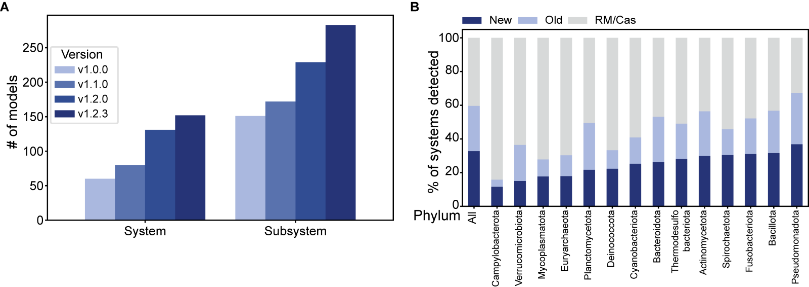
In the ever evolving antiphage defense landscape, we’ve added 92 new systems, now detecting a total of 152. These additions account for more than 33% of the systems detected. Please keep telling us if we are missing some. Full list here: https://defensefinder.mdmlab.fr/wiki/defense-systems
The new web service interface allows anyone to detect defense systems in their preferred genomes and to save previous analysis (without any registration). We also provide genomic visualization and the ability to easily download everything.
One major feedback we had, was the difficulty to understand the hits identified by DefenseFinder and put it in context.
We created a wiki (https://defensefinder.mdmlab.fr/wiki/) where each defense system has its page. Systems found by DefenseFinder are now linked to those wiki-like pages.
The Wiki is usable on its own, allowing research through the diversity of system mechanisms and domains through a searchable table. We added general concepts often used in the field.
It’s a collaborative and participative website, so create missing pages or tell us about them ✍️!

You can find, on the help page of the website, how to contribute. Don’t hesitate to ask us in case of problem.
To allow direct search of the presence of systems in public genomes, we ran DefenseFinder on RefSeq complete genomes database. We provide visualization through interactive tables and graphs that are easily downloadable.
We now live in a world of structure predictions, so we teamed up with Gemma Atkinson’s lab and precomputed all monomers as well as dimers for all known representatives of defense systems. Foldseek results on the AlphaFoldDB and PDB were also precomputed. All these results can be visualized with Mol*, a powerful tool-kit for web-based visualization of protein structures.
Congrats to Florian Tesson, lead author and Rémi Planel for the website development as well as the IT Department of the Institut Pasteur for the technical part.
We organized a hackathon day, internal to the lab, to fill the 152 defense systems’ pages. We also ask expert in the field to write the page on their favorite system. So : huge thanks to everyone who participated in the project, of course MDM Lab members but also wiki contributors : Adi Millman, François Rousset, Daan C. Swarts, Avigail Stokar-Avihail, Nathalie Béchon, Alex L. Gao
We really hope this web service will allow everyone to dive into antiphage defense biology by having easy access to bioinformatics resources.
Don’t hesitate to let us know what you would like to see next and join the fun of creating improving wiki pages!
